Sullivan explores how marine microbiome and virosphere impact oceanic ecosystems
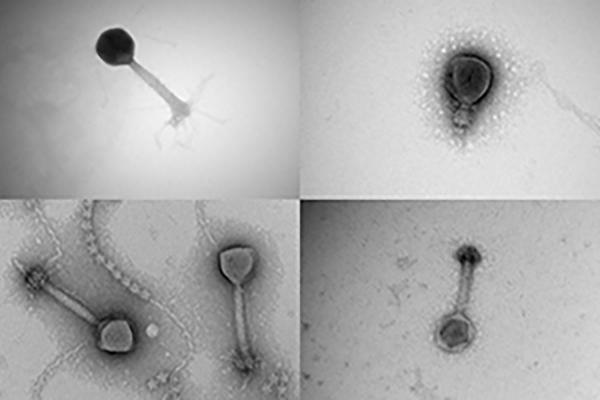
Hover over the four magnifying glasses to view a selection of viruses collected from the Tara Oceans expedition.
The discovery of nearly 200,000 new, marine-dwelling virus species can be traced back to a flash of happenstance — a sheer stroke of good luck.
I

As Sullivan waited to take the stage, he listened to Chris Bowler, a plant and algal biologist, discuss plans to sail around the world collecting samples to capture and catalogue the complex biodiversity of everything in the ocean smaller than fish — from microbial prokaryotes such as bacteria to single-celled eukaryotes like plankton. Bowler’s aim was to provide a more holistic view of the oceanic ecosystem. He wanted to capture everything.
But when Sullivan, then an associate professor of ecology and evolutionary biology at the University of Arizona, stepped in front of the mic to talk about the abundance of viruses present in oceans, Bowler realized his “everything” was missing out on viruses. After Sullivan’s presentation, Bowler sought him out.
“Right there, he said, ‘Do you want to be a part of this?’” Sullivan recalled.
The Tara Oceans Consortium, an expedition of which Bowler is scientific coordinator, launched two years later. The floating laboratory — a 36-meter-long schooner — spent over three years collecting samples from around the world, meticulously filtering seawater through special nets and pumps and filling out the microbiological blind spots overlooked by previous oceanic surveys. As the project’s virus coordinator, Sullivan studied the viruses the expedition collected, and in 2019, he co-authored a study that quantified the viral organisms found and the impact they have on the oceanic ecosystem.
An animated map of the Tara Oceans expedition into the Arctic, which help fill out the catalogue of known marine viral populations.
Sullivan since moved to Ohio State, where he is now a professor in Department of Microbiology and founding director of the recently established Center of Microbiome Science. His expertise lies in the evolution and ecology of phage — or viruses that infect bacteria — with a specialization in marine phages and their interactions within ocean microbiomes. What are these viruses’ role in these tiny networks? How do they change the behavior and biology of their microbial hosts? What unforeseen domino effect does this have on the things we can see, like our oceans, and feel, like climate change?
These are some of the bigger questions Sullivan has. And the answers are found by diving deeper.
“The goal is to understand how viruses and microbes in complex systems work to change the functioning of those ecosystems,” he said. “We haven’t always had the right lens into the biology; a lot of these discoveries in this hidden microbiome space are made when a new tool gets developed and suddenly someone can see. In many ways, it’s just that simple — learning to see.”
Relatively speaking, marine microbiological research is still pretty young, even though it has taken massive steps in recent decades. It was only in 1986 when scientists discovered Prochlorococcus, an aquatic, photosynthetic bacteria. It turns out, Prochlorococcus is responsible for about a quarter of the global photosynthetic production of oxygen and is among the most abundant photosynthetic organisms on the planet.
And until a little over 30 years ago, scientists weren’t even aware of its presence.
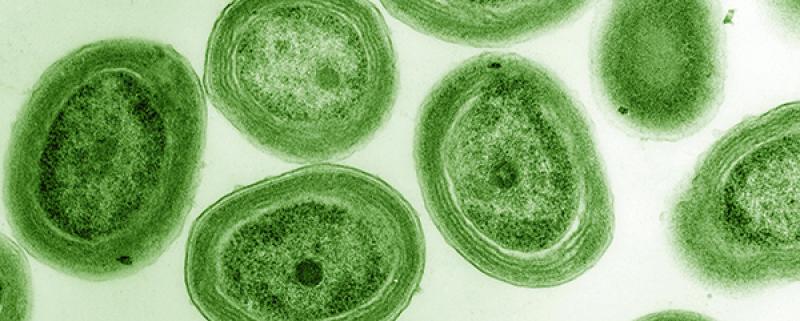
Prochlorococcus, a small cyanobacteria that is believed to be the most abundant photosynthetic organism on Earth.
About a decade after the landmark Prochlorococcus discovery, Sullivan was studying marine science at Long Island University. During an undergraduate research fellowship at the Scripps Institution of Oceanography in San Diego, he was tasked with using phages to label specific, nitrogen-regulated proteins on phytoplankton, or the microscopic plants that populate the oceans.
At the time, all scientists really knew about marine viruses was that there were a lot of them. They didn’t know what they infected or how they interacted with other microbes. Fascinated, Sullivan discovered his calling and decided to pursue his PhD studying viral ecology and evolution.
Since then, Sullivan has contributed to over 100 papers shedding light on what some in the scientific community dub the “virosphere,” the immense, enigmatic world of virus diversity. One recent study examined how viruses steal critical nitrogen-cycling genes in the oceans’ growing oxygen minimum zones, or areas of the ocean where oxygen saturation is disproportionately low due to the disruption of normal gas diffusion and increased carbon dioxide in the atmosphere. Another study explores fundamental viral ecology in these oxygen minimum zones and how viral communities specific to these spaces are structured.
Another study was recently recognized by the International Society for Microbial Ecology (ISME), one of the most prominent associations in the field. The research, which won the inaugural ISME Journal 2020 Best Paper Award, examined how marine phages reprogram their host cells and influence what they release back into an ecosystem, what Sullivan terms their “ecosystem output.” The researchers took the same cells — infected some and left others untouched — and used cutting-edge measurements (whole genome gene expression, protein expression and metabolite responses) to compare their effective impacts on the ecosystem.
The findings belabored how marine viruses influence microbiomes and the ripple effect it has through the ecosystem.
“The viruses reprogram everything about the cell, and not surprisingly, the ecosystem output of virus-infected cells is incredibly different,” Sullivan said. “So, if we think about modelers who are trying to understand the impact of microbes on an ecosystem, they don’t yet think about a virus-infected cell at all. All the models currently capture is the diversity of cell types. What we showed is that the exact same cell type will look entirely different uninfected than when infected with a virus. Thus, these models need to consider not only the diversity of cells, but also their physiological status.”
Sullivan, along with many others at Ohio State, are increasingly aware of how the lessons learned in the oceans translate to microbiomes in other systems, like the human body. To this end, Sullivan established the Center of Microbiome Science in February 2020, and it received official approval in November. Though it opened recently and amid the COVID-19 pandemic, the center already has 75 faculty members across nine colleges and has made significant strides to connect scientists, forge partnerships and promote learning in this fast-moving field.
In many ways, we’re way ahead of the curve here at Ohio State,” Sullivan said. “I’ve been thinking very actively about how to translate this ahead-of-the-curve toolkit broadly across campus. The center was the realization of that idea.”
The center has built out custom microbiome science software tools and data resources that leverage the powerful Ohio Supercomputer Center to comb through immense datasets to better reveal their biological mysteries. To train researchers new to this space, the center has developed a training track of courses designed for students to enter at any level to fill out their understanding and hands-on training in microbiome science. Further, trainee-led working groups in microbiome, virome and advanced ecological statistics are now available to support the growing Ohio State research community in this space, while also providing leadership opportunities for early career researchers.
The center's successes are the result of not just Sullivan, but the broader array of experts who make up the affiliated faculty, including executive director Seth Faith, director of the Infectious Diseases Institute (IDI), and assistant director Vanessa Hale, assistant professor in the College of Veterinary Medicine. With assistance from the (IDI), the Center for Microbiome Science has been poised to make an impression on the microbiome field from the beginning.
“We try to look at the microbiome science landscape and figure out what Ohio State could uniquely and competitively do in these spaces,” Sullivan said. “We started planning in February 2020 when everything looked rosy, and then of course a month later campus and the world were in the midst of the pandemic, and we had to shift tact from in-person faculty and trainee mixers to building out infrastructure and identifying remote learning and interaction opportunities. It’s remarkable what we’ve accomplished since launching.”
Part of what makes the marine microbiome science arena so exciting for Sullivan is its interdisciplinarity — both in terms of who is involved and its outcomes. From microbiologists to atmospheric scientists to environmental engineers, the experts who can stake a claim to the sprawling specialty are vast. And because the techniques used to interrogate the oceanic microbiome can translate across other domains of microbiome science, the impact of their research is limitless.
Sullivan’s oceans toolkit helped Ohio State land $12.5 million from the National Science Foundation to launch a new Biology Integration Institute, which studies how arctic microbes impact climate change and interrogates the roles viruses play in diverse ecosystems. These include ancient and current brines, ancient ice-preserved microbial communities and humans to study smoking, autism, and spinal-cord injury.
“Because the oceans are a complex system, we actually can pretty easily sample them and develop innovative, new technological advancements,” Sullivan said. “These capabilities we’ve developed in the oceans, they’re not being adopted fast enough.”
